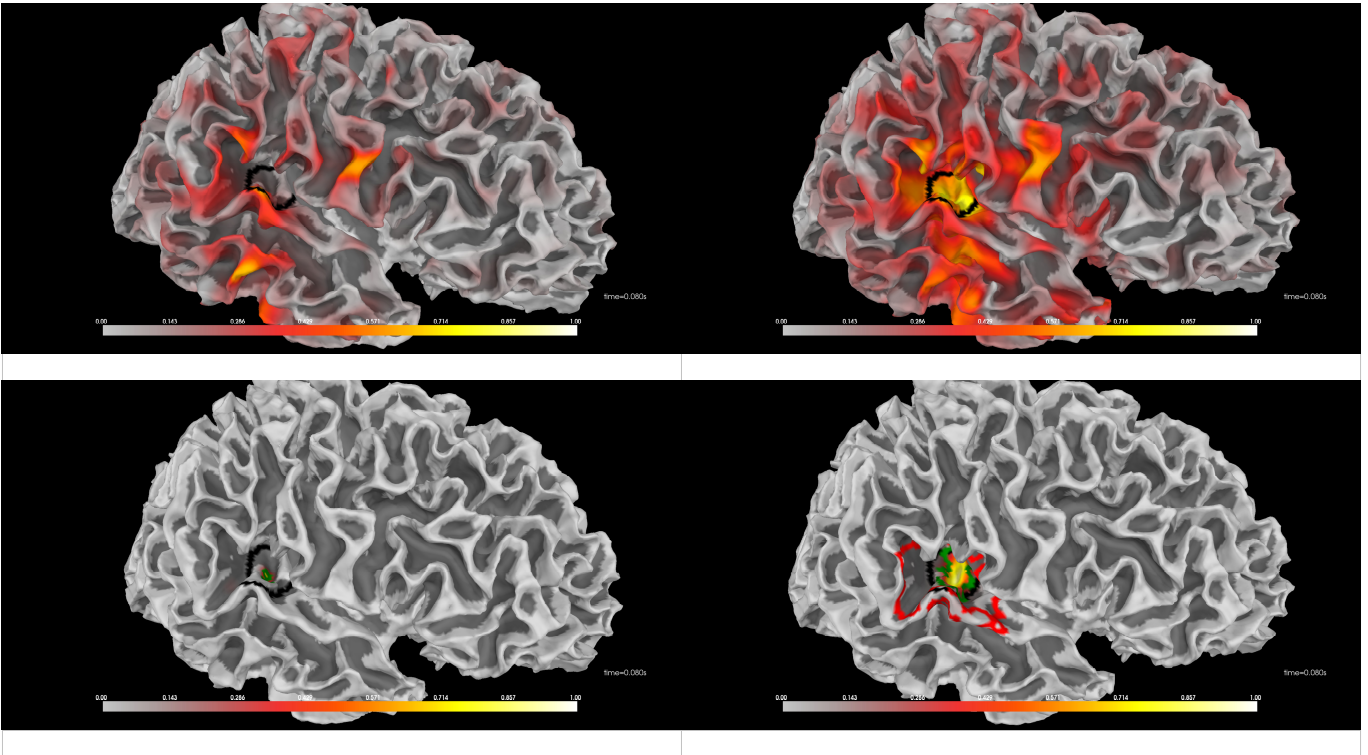
In the context of M/EEG source reconstruction, most distributed source models tend to strongly overestimate the spatial extent of brain activity and underestimate its depth and amplitude (Black contour line: boundary of the “aud-rh” region given by MNE-Python).
The top row shows the results of the MNE and eLORETA algorithms (implemented in MNE-Python with the default parameters), which suffer significantly from these limitations (although eLORETA manages to estimate depth). Sparse Bayesian Learning (SBL, bottom left) underestimates the spatial extent while accurately locating the activity. Combining SBL with spectral graph wavelets, as shown in the bottom right panel, correctly locates the activity (red and green lines are level curves corresponding respectively to 1% and 10% source amplitude levels), estimates its spatial extent and depth, and yields a quantitatively relevant amplitude estimate.
Combining Spatial Wavelets and Sparse Bayesian Learning for Extended Brain Sources Reconstruction
2025. IEEE Transactions on Biomedical Engineering, 1–12. — @HAL
